Enzyme Nomenclature News
Definitions of some common terms used in the field of enzymology
[Prepared by Ron Caspi]
The purpose of this newsletter is to provide a set of accepted definitions for some of the common terms used in the field of enzymology. Over the years, many definitions have been published for these terms. Unfortunately, these definitions have not always been in full agreement with each other, resulting in some confusion. By providing the following definitions, which are endorsed by the Enzyme Commission, the IUBMB hopes to help consolidate the use of these terms.
Enzymes
Enzymes are macromolecular biological catalysts of biochemical reactions. Most enzymes are proteins, but a small number of catalytic RNA molecules, known as ribozymes, are also included in this definition.
Enzymes do not alter the reaction's equilibrium; they accelerate the reaction by lowering its activation energy or by facilitating an alternative catalytic mechanism. Enzymes differ from simple chemical catalysts in showing specificity for individual compounds or groups of compounds.
Generally speaking, enzymes are not consumed during the reaction they catalyse and are ready to catalyse another reaction as soon as they release the reaction's product(s). However, in a small number of cases enzymes may also participate as substrates in the reaction they catalyse (e.g. enzymes that transfer sulfur from an internal iron-sulfur cluster to a substrate), in which case the enzyme must be restored to its active state before it can catalyse an additional cycle.
Proenzymes, apoenzymes and holoenzymes
Some enzymes are produced in an inactive form and require certain modifications before they are able to catalyse reactions. A proenzyme, also known as a zymogen, is an inactive precursor of an enzyme that must be cleaved before becoming active. The cleavage could be performed by the enzyme itself following an environmental change (e.g. in pH) or it may require a peptidase. Sometimes the term preproenzyme is used, indicating that the precursor also contains a signal peptide that directs it to a specific organelle or subcellular localization, which needs to be cleaved to produce the proenzyme. Some enzymes are only active when bound to additional non-protein molecules (see cofactors below). The inactive form of such enzymes is called an apoenzyme, while the active form is called a holoenzyme. These terms are not normally applied to the freely reversible binding of substrates or activators.
Examples for a modification from an apo form to a holo form is the attachment of a 4′-phosphopantetheine group, removed from coenzyme A, to a serine residue of the enzyme, which allows the enzyme to bind acyl moieties.
Reactants, substrates and products
A reactant is a chemical compound that participates in, and is modified during, an enzymic reaction. Compounds that existed prior to the reaction are called substrates, and compounds that are produced during the reaction are called products. Enzymes may act on single or multiple substrates, depending on the specific reaction and enzyme. Following the chemical transformation, the product(s) is/are released from the enzyme.
Cosubstrates
Substrates are sometimes classified further into main substrates and cosubstrates. This distinction derives from the role that the compound plays during the reaction. Cosubstrates commonly provide electrons, protons, phosphate groups, methyl groups etc. for the modification of the main substrate, and most cosubstrates participate in many different reactions and are used by several different enzymes. Since the definition of a cosubstrate depends on its role in a specific reaction, the same compound may be considered a main substrate in one reaction and a cosubstrate in a different one. Examples for common cosubstrates include NADH, S-adenosyl-L-methionine, and ATP.
Cofactors
A cofactor is a non-protein chemical compound that is required by an enzyme for catalysis of a chemical reaction. Cofactors associate with inactive apoenzymes, resulting in formation of active holoenzymes. Although the state of a cofactor may change during the course of the reaction, it remains unchanged at the end of the catalytic cycle and thus, unlike cosubstrates, cofactors are not normally included in the reaction equation. Cofactors can be inorganic ions (e.g. Ca2+), inorganic molecules (e.g. an iron-sulfur cluster) or organic molecules (e.g. pyridoxal 5′-phosphate). In some cases a precursor of the cofactor binds, followed by in situ modification that generates the cofactor. Some cofactors may be formed by modification of existing amino acid residues within the enzyme. Some compounds may act as cofactors for a particular enzyme and a cosubstrate for a different enzyme, depending on whether they are restored to their active state at the end of the reaction.
Prosthetic Groups (not acceptable)
Cofactors that are tightly bound to an enzyme have often been referred to as prosthetic groups. It should be noted that the degree of tightness required for this definition has not been defined, and that the same cofactor may bind differently to different enzymes, making this term imprecise. Thus, it is recommended not to use it.
Coenzymes
The term "coenzyme" has been used in the past to refer to chemical compounds that serve functions that fit both the cosubstrate and cofactor categories. The term is currently included in the IUPAC Gold Book. It is strongly recommended to use the alternative terms cosubstrate and cofactor. For historical reasons, the term is acceptable in the case of the well-established names coenzyme A, B, M, and F420.
[top]
On the nomenclature of fatty acids
[Prepared by Ron Caspi]
The term "fatty acid" was originally coined to describe aliphatic monocarboxylic acids derived from or contained in esterified form in an animal or vegetable fat, oil or wax. However, over the years the term has been expanded to refer to additional, shorter monocarboxylic acids with an aliphatic tail such as propanoic and butanoic acids (but not the shorter formic and acetic acids).
The length of a fatty acid is determined by the length of the longest carbon chain, including the carbon of the carboxy group. Common natural fatty acids usually have an even number of carbons in the longest chain and can be either saturated or unsaturated. In many organisms they are often modified by branching, hydroxylation, methylation, epoxidation and other types of modifications.
Inside living cells fatty acids are rarely found in free form. They are usually bound to coenzyme A or acyl-carrier proteins, or form part of triglycerides, phospholipids, lipopolysaccharides, and cholesterol esters.
Many enzymes that act on fatty acids, as well as the corresponding fatty acyl-CoAs, alcohols, and aldehydes, recognise substrates with a limited range of chain lengths. To classify these enzymes, it is helpful to divide the substrates into smaller groups based on their chain length.
The following subgroups have been defined:
Short-chain fatty acids (SCFA) have 3–5 carbons in the longest chain.
Medium-chain fatty acids (MCFA) have 6–12 carbons in the longest chain.
Long-chain fatty acids (LCFA) have 13–22 carbons in the longest chain.
Very-long-chain fatty acids (VLCFA) have 23–27 carbons in the longest chain.
Ultra-long-chain fatty acids (ULCFA) have more than 27 carbons in the longest chain.
The same terminology applies to fatty acyl-CoAs, fatty acyl-[acyl-carrier proteins], fatty alcohols, and fatty aldehydes.
When used in the accepted names of enzymes, these terms refer to the chain-length range towards which the enzyme is most active. However, it should be noted that in many cases the enzyme may have some activity towards substrates above or below that range.
[top]
Synthases and synthetases
[Prepared by Ron Caspi]
The terms synthase and synthetase, which appear in the accepted names of enzymes, sound similar and have been used interchangeably in the past, yet they have different meanings.
The name synthase is rather generic. Unlike terms such as carboxylase or hydratase, it does not describe a particular type of enzymic activity. All it indicates is that the reaction catalysed by the enzyme is synthetic, rather than catabolic, in nature.
The term is used for different reasons. The most common case is when an enzyme catalyses a complicated transformation that is difficult to define in a concise way. An example for such use is EC 1.13.11.79, aerobic 5,6-dimethylbenzimidazole synthase. This enzyme catalyses the fragmentation and contraction of a bound reduced flavin mononucleotide (FMNH2) and cleavage of its ribityl tail to form 5,6-dimethylbenzimidazole and D-erythrose 4-phosphate in a reaction that has been described as "cannibalization" [1].
A second case where the term synthase is used is when the enzyme's activity involves two or more reactions of different nature, making it difficult to decide which of the activities should be used for naming the enzyme. For example, EC 4.2.1.20, tryptophan synthase, catalyses a lyase reaction that splits 1-C-(indol-3-yl)glycerol 3-phosphate into indole and D-glyceraldehyde 3-phosphate, followed by a transfer reaction, transferring the indole to an L-serine, replacing its hydroxyl moiety and forming L-tryptophan.
The third reason to use the term synthase is simply to preserve names that have been used for some time and adapted by the scientific community. Since the term is not incorrect, it is sometime preferable to keep it even though a more precise name is feasible. An example is EC 1.1.1.318, eugenol synthase, which could have been called by the more precise name coniferyl ester reductase.
In addition to the accepted names, the Enzyme Commission also specifies systematic names that provide a more precise description of the reaction. In the case of the three enzymes mentioned above, the corresponding systematic names are FMNH2 oxidoreductase (5,6-dimethylbenzimidazole-forming); L-serine hydro-lyase [adding 1-C-(indol-3-yl)glycerol 3-phosphate, L-tryptophan and D-glyceraldehyde-3-phosphate-forming]; and eugenol:NADP+ oxidoreductase (coniferyl ester reducing). As is evident, the systematic names are much more complicated in these cases, explaining why the term synthase has been so popular. As of August 2019, 848 Enzyme Commission entries use that term in their accepted name.
The term synthetase, on the other hand, is rather specific. Its proper use has been reserved only for ligases, enzymes that catalyse the joining together of two molecules, powered by the hydrolysis of a diphosphate bond in ATP or a similar triphosphate (classified under class 6). Acknowledging the confusion that the two similar terms have caused, the Enzyme Commission decided in 1983 to abandon the use of the term synthetase for accepted names, replacing it with names of the type X—Y ligase. Most of the names have been subsequently modified to reflect this decision. However, a small number of cases where the reaction is more complex (for example, an intramolecular ligation as in EC 6.1.3.1, olefin β-lactone synthetase) have retained the term synthetase in their accepted names. As of August 2019, only 7 such entries remain.
1. Taga, M.E., Larsen, N.A., Howard-Jones, A.R., Walsh, C.T. and Walker, G.C. BluB cannibalizes flavin to form the lower ligand of vitamin B12. Nature 446:449 (2007).
[top]
Ribonuclease classification
[Prepared by Keith Tipton]
The classification of the ribonucleases has been a matter of some contention. Whereas some appear to act as simple hydrolases, cleaving the 3′,5′-phosphatodiester bonds linking adjacent nucleotides, others, such as pancreatic ribonuclease, cleave to give a 2′,3′-cyclophosphate derivative, which is subsequently hydrolysed to the 3′-phosphate, as shown below.
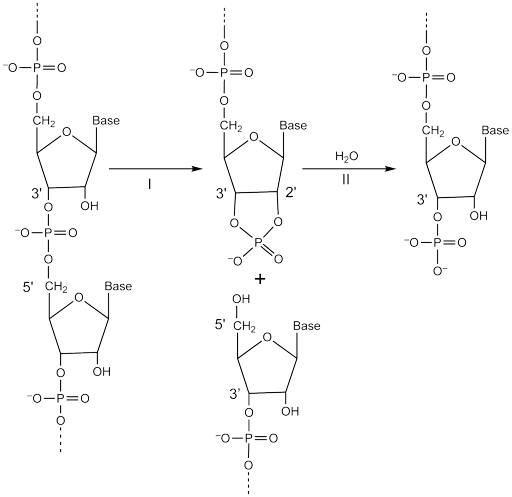
Pancreatic ribonuclease was listed among those enzymes transferring phosphate groups, Enzymes catalysing other phosphate-transfer reactions, as entry 502 in the Table of Enzymes published by Dixon & Webb in 1958 [1]. When the Enzyme List was formally divided into specific reaction classes, it was listed as a transferase, (EC 2.7.7.16) in 1961 [2], but, since the overall reaction was hydrolysis, it was reclassified as a hydrolase (EC 3.1.27.5) in 1972 [3]. However, it became clear that neither of these classification types adequately described the activity of this enzyme. The cyclophosphate produced in reaction I is actually released from the enzyme and the subsequent hydrolytic step (reaction II), which occurs by a reversal of reaction I with the hydroxyl group of water replacing the 5′-hydroxyl group of ribose, only occurring to any appreciable extent when the formation of the cyclophosphate product is essentially complete. Thus the reaction may be regarded as that of a lyase (a phosphorous-oxygen lyase) since it ‘cleaves bonds by means other than by hydrolysis or oxidation’. This is the class of enzyme where two (or more) substrates are involved in one reaction direction, but there is one compound fewer in the other direction and when acting on the single substrate, a molecule is eliminated and this generates either a new double bond or, in this case, a new ring structure.
Because of this, pancreatic ribonuclease and some other ribonucleases have now been reclassified as phosphorous-oxygen lyases (EC 4.6.1.-). Those ribonucleases which appear to behave as simple hydrolases remain in EC 3 along with the DNases, which cannot form 2′,3′-cyclophosphate products.
1. Dixon M. & Webb EC. (1958) Enzymes, Longmans Green, London, pp. 185–227.
2. Report of the Commission on Enzymes of the International Union of Biochemistry (1961). Pergamon Press, New York & London.
3. Enzyme Nomenclature (1973) Recommendations (1972) of the Commission on Biochemical Nomenclature on the Nomenclature and Classification of Enzymes, Together with their Units and the Symbols of Enzyme Kinetics. Elsevier, Amsterdam.
[top]
Translocases (EC 7): A new EC Class
[Prepared by Keith Tipton]
Six enzyme classes have been recognized since the first Enzyme classification and nomenclature list was first approved by the International Union of Biochemistry in 1961. These were based on the type of reaction catalysed: Oxidoreductases (EC 1), Transferases (EC 2), Hydrolases (EC 3), Lyases (EC 4), Isomerases (EC 5) and Ligases (EC 6). However, it has become apparent that none of these could describe the important group of enzymes that catalyse the movement of ions or molecules across membranes or their separation within membranes. Several of these involve the hydrolysis of ATP and had been previously classified as ATPases (EC 3.6.3.-), although the hydrolytic reaction is not their primary function.
These enzymes have now been classified under a new EC class of translocases (EC 7).The reactions catalysed are designated as transfers from 'side 1′ to 'side 2′ because the designations ‘in’ and ‘out’ (or ‘cis’ and ‘trans’), which had been used previously, lack clarity and can be ambiguous. The comments associated with each entry then describe the specific translocations catalysed.
The subclasses designate the types of ion or molecule translocated:
EC 7.1 contains enzymes catalysing the translocation of hydrons (hydron being the general name for H+ in its natural abundance),
EC 7.2 contains those catalysing the translocation of inorganic cations and their chelates,
EC 7.3 contains those catalysing the translocation of inorganic anions,
EC 7.4 contains those catalysing the translocation of amino acids and peptides,
EC 7.5 contains those catalysing the translocation of carbohydrates and their derivatives
EC 7.6 contains those catalysing the translocation of other compounds.
The sub-subclasses concern the reaction that provided the driving force for the translocation, where these are relevant:
EC 7.x.1 translocations linked to oxidoreductase reactions
EC 7.x.2 translocations linked to the hydrolysis of a nucleoside triphosphate
EC 7.x.3 translocations linked to the hydrolysis of a diphosphate
EC 7.x.4 translocations linked to a decarboxylation reaction
Exchange transporters that are not dependent on enzyme-catalysed reactions, such as the exchange of ions across membranes, are not included and pores that change conformation between open and closed states in response to phosphorylation or some other catalysed reaction are classified under EC 5.6 (Macromolecular conformational isomerases).
(We are grateful to Dr Amanda Mackie (Macquarie University) for her advice on the content of this EC class)
[top]
How to name atoms in phosphates, polyphosphates, their derivatives and mimics, and transition state analogues for enzyme-catalysed phosphoryl transfer reactions (IUPAC Recommendations 2016)
[Prepared by Gerry Moss]
The Protein Databank (PDB) includes many examples of phosphoryl transferase enzymes (EC 2.7) that contain phosphates or polyphosphates and their derivatives or mimics. A new IUPAC Recommendation published in Pure and Applied Chemistry* recommends how to identify and uniquely identify each atom of the phosphate or polyphosphate.
For monoesters the phosphorus atoms are identified as PA, PB, PG, PD, etc., starting from the ester end. For diesters the new recommendations explain how to determine which ester should be used to number the phosphorus atoms. When both a nucleic acid and a non-nucleic acid groups are present, the former takes precedence. If both are nucleic acid groups, then alphabetic order (A > C > G > T > U) is used.
With phosphate multi-esters, the phosphorus atoms are identified by the position at which they are attached. For example, in fructose 1,6-bisphosphate the two phosphorus atoms are named P1 and P6. In polyphosphates the phosphorus atoms are named PA1, PB1, PG1, etc.
The oxygen atom directly bonded to the sugar moiety is numbered using the number of the carbon it is attached to. For example, in ATP it is O5′. The other three oxygen atoms of PA are named O1A, O2A and O3A, where O3A bonds to PB, O1A is pro-R, and O2A is pro-S.
When a terminal phosphoryl oxygen is replaced by sulfur, fluorine or nitrogen, the remaining two oxygen atoms are prochiral and are identified accordingly. For example, for ATP with a sulfur substitution, the sulfur atom is named S1G and the two oxygens are O2G (pro-R) and O3G (pro-S).
Where the oxygen is hydrogen bonded to an adjacent amino acid (with bond length 3 Å), the primary sequence number of the amino acid is used to determine the oxygen numbering, as in O1A, O2A, O3A, etc.
Other topics discussed in this recommendation include substitution by other atoms, trigonal bipyramidal phosphate transition state analogues, and metal coordination.
*Pure Applied Chem. 2017, 89(5), 653–675 DOI 10.1515/pac-2016-0202.
[top]
Classification of cytochrome P-450 enzymes by the Enzyme Commission
[Prepared by Ron Caspi]
Cytochrome P-450 enzymes (CYPs) are a large family of heme-containing proteins found in all kingdoms of life. The term "P-450" is derived from the spectrophotometric peak at the wavelength of the absorption maximum of the enzyme (450 nm) when it is in the reduced state and complexed with carbon monoxide. More than 50,000 distinct CYP proteins have been described.
The CYPs are classified into several groups, the main ones being eukaryotic microsomal enzymes, eukaryotic mitochondrial enzymes, and bacterial enzymes. While most CYPs require electrons that originate from NADPH, almost no such enzyme can receive the electrons directly from NADPH, and a secondary electron transfer protein is involved. The classification of CYPs by the Enzyme Commission is determined by the type of reaction they catalyse and the type of electron donor with which they interact.
For microsomal enzymes, the usual electron donor is EC 1.6.2.4, NADPH-hemoprotein reductase. When the reaction involves monooxygenation and formation of a single molecule of water, the enzyme should be classified under EC 1.14.14: oxidoreductases acting on paired donors, with incorporation or reduction of molecular oxygen, with reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen into the other donor (e.g. EC 1.14.14.16, steroid 21-monooxygenase).
Bacterial CYPs usually utilize ferredoxin. These enzymes are classified under EC 1.14.15: oxidoreductases acting on paired donors, with incorporation or reduction of molecular oxygen, with reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen into the other donor (e.g. EC 1.14.15.8, steroid 15β-monooxygenase).Mitochondrial CYPs utilize a specialized ferredoxin, known as adrenodoxin, as their electron donor, and are thus also classified under EC 1.14.15. (e.g. EC 1.14.15.15, cholestanetriol 26-monooxygenase).
When the reaction involves the formation of two molecules of water, the enzyme should be classified under EC 1.14.19: oxidoreductases acting on paired donors, with incorporation or reduction of molecular oxygen, with oxidation of a pair of donors resulting in the reduction of O2 to two molecules of water (e.g. EC 1.14.19.52, camalexin synthase).
Exceptions do occur — for example, CYPs from the CYP74 family catalyse dehydration reactions that do not require oxygen or an electron donor and are classified under EC 4.2.1 (e.g. EC 4.2.1.121, colneleate synthase). A number of CYPs catalyse an isomerization reaction and should be classified under the proper sub-subclass for the particular isomerization reaction (e.g. EC 5.3.99.4, prostaglandin-I synthase).
Unfortunately, in the past many CYPs have been misclassified under EC 1.14.13, which should be used for enzymes that use NADH/NADPH as their direct electron source. In the coming months these entries will be reclassified under the proper sub-subclass.
[top]
Archaeal enzymes
[Prepared by Ida Schomburg]
Besides the eukaryotes and the prokaryotes, the archaea represent the third domain of life. They are unicellular and similar to bacteria in shape and size, but their metabolism resembles eukaryotic behaviour in many aspects, notably the enzymes involved in archaeal DNA replication. Archaea are ubiquitous and have been found in practically all environments. Species living under extreme conditions such as hot, highly saline, alkaline or acidic waters, or under high pressure in the deep sea, were the first to be discovered. These "extremophiles" developed special strategies to maintain the stability of their cellular structure and to thrive despite limited nutrient supply, making for unique and interesting enzymes. In this short communication we will list of a few of the specialized enzymes found in archaea.
For surviving under harsh conditions the cellular structures must be highly stable. Archaeal membranes achieve that by containing archaetidylserine (2,3-bis-(O-phytanyl)-sn-glycero-1-phospho-L-serine), which is based on glycerol ether components instead of glycerol fatty acid esters. The enzymes involved in the biosynthesis of archaeal lipids have been newly classified and are shown in the diagram for archaetidylserine biosynthesis.
Polyamines are required to maintain the stability of double-stranded DNA. Where eukaryotes and prokaryotes produce linear spermine and spermidine, archaea synthesize branched-chain and long-chain polyamines, such as N4-bis(aminopropyl)spermidine or caldopentamine, which confer a higher stability. The enzymes involved in their synthesis are classified as EC 2.5.1.127 and EC 2.5.1.128.
During their long evolution archaea developed strategies for taking up nutrients very efficiently. The substrate specificity of archaeal enzymes is often less strict than that of their bacterial counterparts. For example, two new enzymes of the archaeal Entner-Doudoroff pathway have been described, one being able to dehydrogenate either glucose or galactose (EC 1.1.1.360) and the other, with even less specificity, is able to dehydrogenate a broad range of aldoses (EC 1.1.1.359). In addition, archaea do not follow the classical route of the Entner-Doudoroff pathway, and instead use semi- or non-phosphorylative variants. Three enzymes that participate in these pathways have been newly classified: EC 1.2.1.89, EC 1.2.99.8 and EC 4.1.2.55.
[top]
Fatty acid desaturases
[Prepared by Ron Caspi]
Fatty acid desaturases are enzymes that convert a single bond between two carbon atoms in a fatty acyl chain to a double bond. A common property for all known desaturases is their requirement for molecular oxygen and a source of electrons.
The classification of desaturase enzymes is complicated by the fact that they can be classified by several criteria. First, they differ in their substrate preferences. Free fatty acids are very rare in the cell, and fatty acids are usually found bound to either coenzyme A, acyl-carrier proteins, or as part of a glycerolipid. Desaturase enzymes are specific for one of these forms, and often are also specific for a substrate of a particular size or size range. Next, the enzymes differ in their electron donor. Most fatty acid desaturases utilize electrons provided by either ferredoxin or cytochrome b5, although some exceptions do occur. In many of the cytochrome b5-dependent enzymes gene fusion has resulted in a cytochrome b5 domain integrated into the enzyme. The enzymes can also be classified based on the position of the double bond that they introduce - some introduce it at a certain distance from the carboxylic end of the fatty acid, others determine the position based on its distance from the methyl end of the substrate, and yet others introduce the new double bond only at a particular position relative to an existing double bond. Finally, desaturases differ in the type of bond they introduce mdash some introduce a cis (Z) bond, others introduce a trans (E) bond, yet some introduce a mixture of both types.
Owing to the rapid increase in the number of different desaturases that were discovered in bacteria, algae, plants, fungi, and animals, the existing classifications in the Enzyme List became unsatisfactory. We have recently reviewed the existing literature and updated the list, resulting in a major revision of the fatty acid desaturase entries. We have revised most of our existing entries to eliminate ambiguity and added a large number of new entries, achieving close to full coverage of current knowledge in the field.
We tried to address all of the considerations described above. For example, EC 1.14.19.35 stands for sn-2 acyl-lipid ω-3 desaturase (ferredoxin), an enzyme that acts on acyl chains attached to position sn-2 of glycerolipids, introduces a double bond three carbons away from the methyl end of the chain, and accepts electrons from ferredoxin. Naturally, additional information can be gathered by looking at the reaction equations and reading the comment and references.
To see the entries, search the list for the name "desaturase" and scroll down to the entries that start with 1.14.19.
[top]
